Details
-
Type:
Story
-
Status: In Progress
-
Priority:
Major
-
Resolution: Unresolved
-
Affects Version/s: 2.11.0, 2.11.5.0, 2.11.4
-
Fix Version/s: 2.11.1
-
Component/s: annotation, Structures
-
Labels:
Description
Hi,
I'm a structural biologist, and I find Jalview extremely useful for making annotated protein sequence alignments!
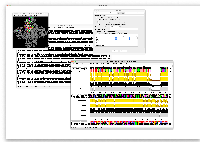
There is one feature that I would really love though: I would like to be able to have the upper scale ignore gaps. That is, to have the upper scale reflect the sequence numbering of a specified reference sequence, rather than the alignment position. This is very useful when cross-referencing with a structure, so I can quickly check from an alignment figure which particularly interesting residue is which numbering in a structure.
Currently I do this manually in a vector graphics program (see below), which becomes rather tedious when one is dealing with an alignment of 5000 residue protein sequences :)
The only other thing I would love is the ability to add custom, editable secondary structure annotations at the bottom of the alignment, although I realize that might be a bit of a tricky one... I'd love to be able to highlight a region of sequence and define as beta sheet, alpha helix, coil or disordered via a context menu and then have that appear as an annotation layer below the sequence.
Keep up the good work, Jalview is a wonderful and essential piece of software!
Cheers
Oliver Clarke (Columbia University, NY)
PS: (Wasn't sure where to post this - I tried posting to jalview-discuss but I don't think it went through.)
I'm a structural biologist, and I find Jalview extremely useful for making annotated protein sequence alignments!
There is one feature that I would really love though: I would like to be able to have the upper scale ignore gaps. That is, to have the upper scale reflect the sequence numbering of a specified reference sequence, rather than the alignment position. This is very useful when cross-referencing with a structure, so I can quickly check from an alignment figure which particularly interesting residue is which numbering in a structure.
Currently I do this manually in a vector graphics program (see below), which becomes rather tedious when one is dealing with an alignment of 5000 residue protein sequences :)
The only other thing I would love is the ability to add custom, editable secondary structure annotations at the bottom of the alignment, although I realize that might be a bit of a tricky one... I'd love to be able to highlight a region of sequence and define as beta sheet, alpha helix, coil or disordered via a context menu and then have that appear as an annotation layer below the sequence.
Keep up the good work, Jalview is a wonderful and essential piece of software!
Cheers
Oliver Clarke (Columbia University, NY)
PS: (Wasn't sure where to post this - I tried posting to jalview-discuss but I don't think it went through.)