Details
-
Type:
New Feature
-
Status: Closed
-
Priority:
Major
-
Resolution: Fixed
-
Affects Version/s: None
-
Component/s: sequencefeatures
-
Labels:
-
Epic Link:
Description
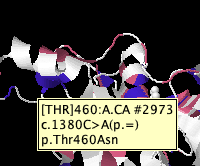
Ambition: enable feature visualisation on CDS / protein / structure, as tailored (features, colours, filters) in the 'native' alignment of the feature.
- add an option to 'Show CDS features' on peptide (and vice versa?)
- locate and display 'complement' features on the fly via sequence mappings
Attachments
Issue Links
- blocks
-
JAL-3304 Export linked features
-
- Closed
-
-
JAL-3535 View specific feature settings dialog title shows name of associated view
-
- Closed
-
- related with
-
JAL-3534 Multiple feature settings dialogs can be opened for a view
-
- Closed
-
-
JAL-3533 Remember positioning of Feature Settings dialog
-
- Closed
-
-
JAL-3684 Cancel feature colour change in split frame doesn't update colour swatch
-
- Open
-
-
JAL-3536 Can't show cross-refs twice for aligned transcripts + reference locus retrieved from Ensembl
-
- Closed
-
-
JAL-3567 Genomic position shown in virtual feature tooltip
-
- Closed
-
-
JAL-3613 Peptide-to-CDS tracking broken for EMBL gene products
-
- Closed
-
-
JAL-3303 Informative tooltip on structure residue for Jalview feature(s)
-
- In Progress
-
-
JAL-3302 Transfer 'virtual' features to Chimera
-
- Closed
-
-
JAL-3564 Allow 'virtual CDS/Protein' features to be overlaid without opening a CDS/Protein linked alignment
-
- Open
-
-
JAL-3251 Add biotype info to sequence mappings
-
- In Progress
-
-
JAL-2792 Provide tabular view of (one) sequence feature's details
-
- Closed
-