Details
-
Type:
New Feature
-
Status: Resolved
-
Priority:
Major
-
Resolution: Fixed
-
Affects Version/s: 2.7
-
Fix Version/s: 2.8
-
Component/s: DAS, data retrieval services, Feature Request
-
Labels:
-
Environment:For Windows 7
Description
It would be very useful to be able to capture in a CSV or Excel format file all of the information gathered on highlighted sequences, when using the View => Feature Settings... => DAS Settings tab.
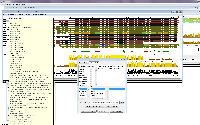
At the moment, all of the output information is only visible by mousing over the sequences for which this data was gathered using the DAS retrieval feature in Jalview 2.7, and even there are some icons indicating links to the sources of such information appearing on the window, but no direct links appear, nor does it appear that there is any easy function integrated in Jalview to create an output file with such information. I'm told by Jim Procter that it's not a hard thing to achieve, and indeed finding a way of doing so would facilitate enormously my work as a curator, where I would be able to easily retrieve all this information as a result of me performing a multiple sequence alignment with my sequences of interest.
Would it be possible to create such a feature in Jalview 2.7?
I had spoken about this with Jim at the Jalview course in Hinxton this early December 2011 (see attached image to show what I am referring to).
Many thanks,
Carolina
At the moment, all of the output information is only visible by mousing over the sequences for which this data was gathered using the DAS retrieval feature in Jalview 2.7, and even there are some icons indicating links to the sources of such information appearing on the window, but no direct links appear, nor does it appear that there is any easy function integrated in Jalview to create an output file with such information. I'm told by Jim Procter that it's not a hard thing to achieve, and indeed finding a way of doing so would facilitate enormously my work as a curator, where I would be able to easily retrieve all this information as a result of me performing a multiple sequence alignment with my sequences of interest.
Would it be possible to create such a feature in Jalview 2.7?
I had spoken about this with Jim at the Jalview course in Hinxton this early December 2011 (see attached image to show what I am referring to).
Many thanks,
Carolina