Details
-
Type:
Bug
-
Status: Closed
-
Priority:
Major
-
Resolution: Fixed
-
Affects Version/s: 2.11.2.6
-
Fix Version/s: 2.11.3.0
-
Component/s: awt-gui, file format issue
-
Labels:
-
Environment:Slackware linux
Description
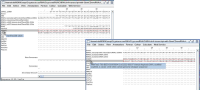
After converting a fasta alignment (right section of image "no-description..jpg")) to the stockholm format (left section) the sequence descriptions (see mouseovers) are lost.
This is very unhelpful as in the sequence description accession numbers and nucleotide numbers can be stored, including the full species name. When losing this information alignments cannot be reproduced.
Sequence information in the stockholm format (examples used as on https://en.wikipedia.org/wiki/Stockholm_format) can be stored with a line like:
#=GS <name> DE <description> for example:
#=GS VanFra/258-1026 DE BEDY01000003.1:1711800-1712595 Vanrija fragicola JCM 1530
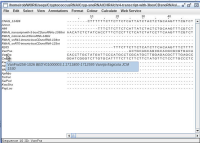
Adding this comment in a text-editor to the stockholm file and then reloading this in jalview works (see "with manually-added...jpg").
This work-around is useless as the annotation is lost after saving this alignment from jalview.
Included annotations for secondary structure information (#=GC SS_cons or #=GR <name> SS) are maintained though.
So in principle it could be expected that other stockholm notes would be kept as well.
This is very unhelpful as in the sequence description accession numbers and nucleotide numbers can be stored, including the full species name. When losing this information alignments cannot be reproduced.
Sequence information in the stockholm format (examples used as on https://en.wikipedia.org/wiki/Stockholm_format) can be stored with a line like:
#=GS <name> DE <description> for example:
#=GS VanFra/258-1026 DE BEDY01000003.1:1711800-1712595 Vanrija fragicola JCM 1530
Adding this comment in a text-editor to the stockholm file and then reloading this in jalview works (see "with manually-added...jpg").
This work-around is useless as the annotation is lost after saving this alignment from jalview.
Included annotations for secondary structure information (#=GC SS_cons or #=GR <name> SS) are maintained though.
So in principle it could be expected that other stockholm notes would be kept as well.
Attachments
Issue Links
- related with
-
JAL-4179 Common nucleotide symbols turns sequence into 'protein', cannot set colour to 'nucleotide'
-
- Open
-